 Collaborators: David Belnap, PhD, Peter Shen, PhD
Collaborators: David Belnap, PhD, Peter Shen, PhD
Department: Biochemistry, Electron Microscopy Core
Project
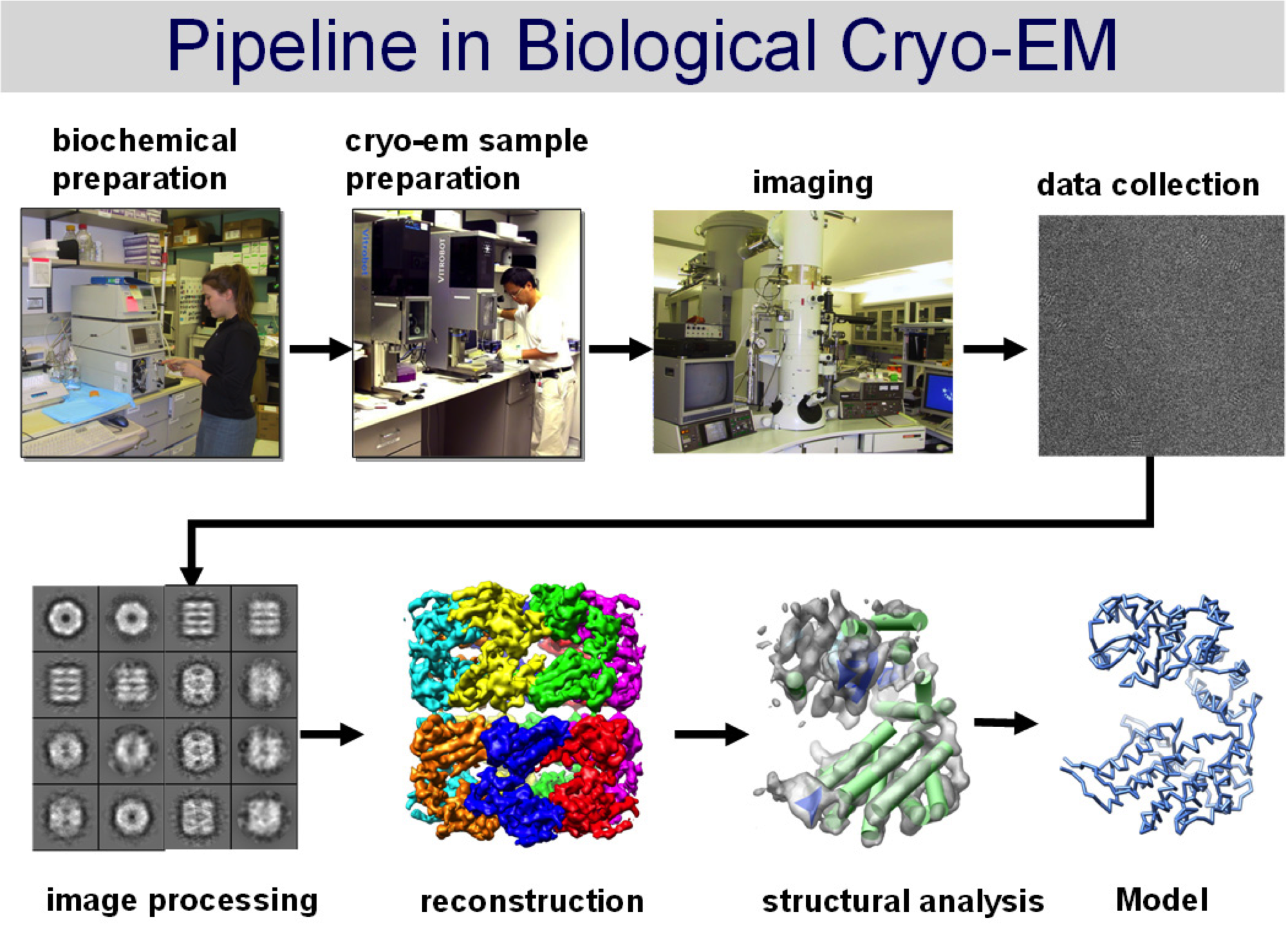
We are on the horizon of a new era for structural biology. Pioneering efforts in both the academic and the private sector led to the invention and commercialization of direct electron detectors for use in electron cryo-microscopy (cryoEM). In the last two years these detectors have enabled 3D structure determination of biological complexes at near-atomic resolution complexes which have been recalcitrant or impossible to study by more mature structural methods like x-ray crystallography or magnetic resonance spectroscopy. University of Utah investigators and administrators alike are committed to bringing this technology to our campus and prepared to realize its full potential.
Two features of CMOS-based direct electron detectors have enabled near-atomic resolution structure determination by cryoEM: 1) improved Detective Quantum Efficiency (DQE), and 2) much faster read-out rates. Together, these innovations have alleviated both of the main problems that limited the resolution of cryoEM studies in the past: 1) the loss of contrast due to low efficiency detectors; and 2) the image distortions caused by electron beam-induced movements of the sample. The enhanced DQE leads to increased signal-to-noise in the images themselves, and this improves the accuracy of both particle detection and orientation determination. Coupled with fast read-out rates, it is now possible to record movies rather than still shots of a sample and to accurately track and correct computationally the movement of a sample as the electron beam passes through it. These advances have made it possible to resolve atomic resolution features of true single particle without crystallization, to obtain atomic-resolution electron diffraction data from microscopic three-dimensional crystals that are far too small for analysis by x-ray diffraction, and to recover molecular-scale features from whole-cell electron tomograms.
CryoEM data processing requires us to split massive datasets into many smaller subsets before distributing the computational burden over a cluster of many Central Processing Units (CPUs). In order to fully capitalize upon the opportunities presented by the newly-developed direct electron detectors, however, many users have moved away from CPU-architectures to Graphics Processing Units (GPUs). GPUs, unlike CPUs, have hundreds of processing cores integrated into one unit and are designed to perform massively parallelized computing by applying single instructions to multiple datapoints (e.g. pixels or whole images). For cryoEM, the use of GPUs for data processing has led to significant speed increases for many of the discrete stages required during 3D reconstruction.
BIDAC Contacts: Josh Cates, Clement Vachet
BIDAC Expertise
Adapt and implement existing CUDA-code to our microscope and image processing pipeline.